Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid
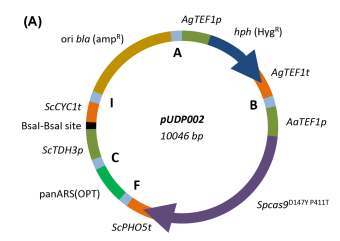
Abstract: While CRISPR-Cas9-mediated genome editing has transformed yeast research, current plasmids and cassettes for Cas9 and guide-RNA expression are species specific. CRISPR tools that function in multiple yeast species could contribute to the intensifying research on non-conventional yeasts. A plasmid carrying a pangenomic origin of replication and two constitutive expression cassettes for Cas9 and ribozyme-flanked gRNAs was constructed. Its functionality was tested by analyzing inactivation of the ADE2 gene in four yeast species. In two Kluyveromyces species, near-perfect targeting (>= 96%) and homologous repair (HR) were observed in at least 24% of transformants. In two Ogataea species, Ade(-) mutants were not observed directly after transformation, but prolonged incubation of transformed cells resulted in targeting efficiencies of 9% to 63% mediated by non-homologous end joining (NHEJ). In an Ogataea parapolymorpha ku80 mutant, deletion of OpADE2 mediated by HR was achieved, albeit at low efficiencies (< 1%). Furthermore the expression of a dual polycistronic gRNA array enabled simultaneous interruption of OpADE2 and OpYNR1 demonstrating flexibility of ribozyme-flanked gRNA design for multiplexing. While prevalence of NHEJ prevented HR-mediated editing in Ogataea, such targeted editing was possible in Kluyveromyces. This broad-host-range CRISPR/gRNA system may contribute to exploration of Cas9-mediated genome editing in other Saccharomycotina yeasts.
Author(s): Juergens, H; Varela, JA; de Vries, ARG; Perli, T; Gast, VJM; Gyurchev, NY; Rajkumar, AS; Mans, R; Pronk, JT; Morrissey, JP
FEMS YEAST RESEARCH
Volume: 18 Published: MAY 2018
DOI: 10.1093/femsyr/foy012